QligFEP is a best-in-class FEP engine, combining excellent accuracy with superior computational performance.

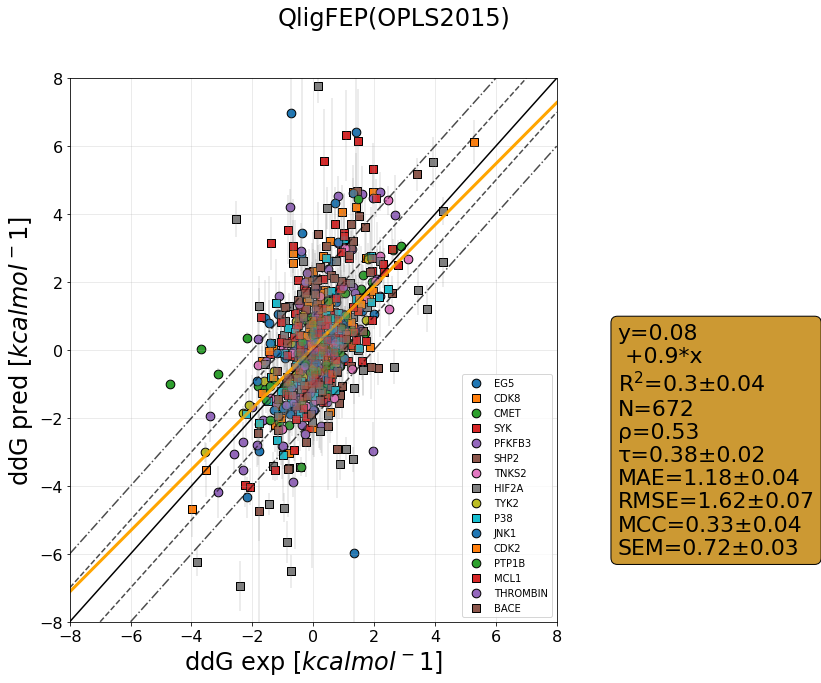
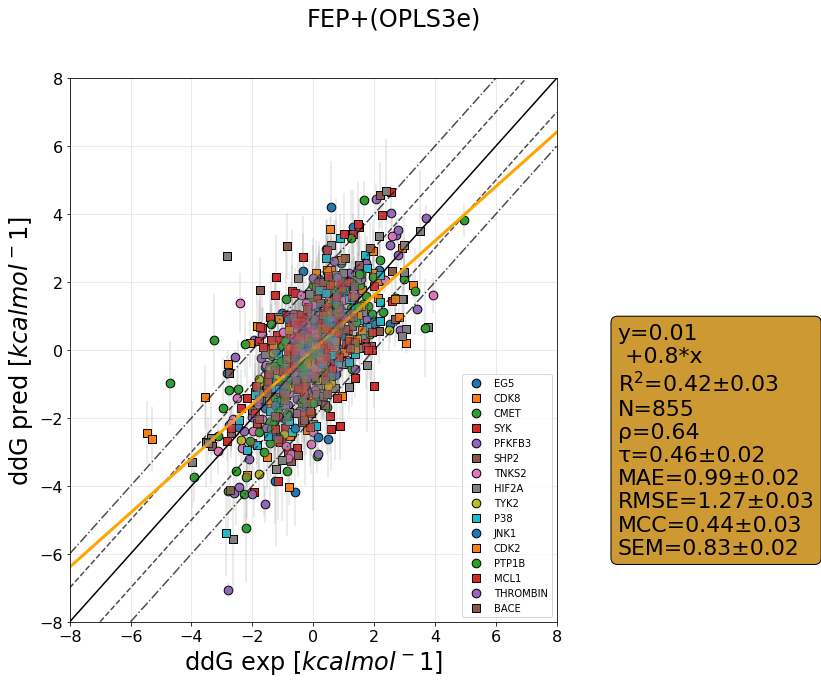
QligFEP has recently been benchmarked on a large dataset consisting of 16 protein targets, accumulating data for over 400 ligands of broad chemical families. The benchmark dataset is a collection of the Wang dataset, published in 2015, together with the recent Merck dataset, which we have run with three different forcefields: OPLS2015, openFF1.0, and the recently released openFF2.0.
The results are shown as cycle closure corrected relative binding free energies for each edge, generated for each target using our propietary QmapFEP sampling engine. QligFEP is impressively accurate with an average mean unassigned close to 1 kcal/mol over all targets, which is equivalent to other methods using proprietary forcefields (FEP+, included for comparison). On some targets, QligFEP significantly outperforms this and other approaches.
Moreover, we analyzed the computing time required for each dataset, which is well below any other FEP engine on the market. The reasons are to be found in the optimized QmapFEP engine (note the number of edges to cover the dataset as compared to e.g. FEP+ in the graphs below), in combination with the spherical boundary conditions and optimized sampling described in our original report. The combined use of QligFEP and QmapFEP shows itself as a very powerful tool for drug discovery programs.
Contact us to get a price estimate to run an FEP campaign on your target.