A one-stop package for the optimized modeling and molecular dynamics simulation of GPCRs.
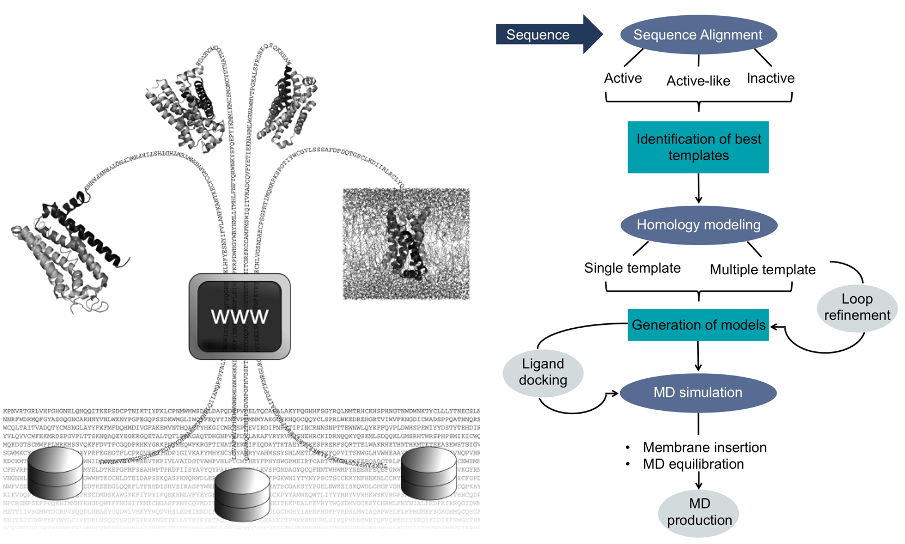
GPCR-ModSim is a centralized and easy to use service dedicated to the structural modeling of G Protein-Coupled Receptors (GPCRs). 3D molecular models can be generated from amino acid sequence by homology-modeling techniques, considering different receptor conformations.
GPCR-ModSim includes PyMemdyn, a stand-alone membrane insertion and molecular dynamics (MD) equilibration protocol, which can be used to refine the generated model or any GPCR structure.
GPCR ModSim was the best automated protocol to model the structure of the 5-HT2b serotonin receptor in the GPCRdock competition.
GPCR-ModSim: A comprehensive web based solution for modeling G-protein coupled receptors
M. Esguerra, A. Siretskiy, X. Bello, J. Sallander, H. Gutiérrez-de-Terán, Nucleic Acids Research 2016, 44 (W1), W455–W462.