In silico mutagenesis with free energy perturbation (FEP) simulations.

QresFEP is based on a smooth annihilation protocol to perturb the aminoacid sidechain of interest. This protocol showed high convergence and robustness.
The approach has been tested on multiple targets to estimate the effect of single point mutations on protein-ligand binding affinity, thermal stability or conformational equilibrium. QresFEP showed a consistent strong signal for predicting these effects.
QresFEP is a flexible approach which can handle different forcefields and sampling options. The approach is fast and accurate, owing to the spherical boundary MD simulations within our Q package.
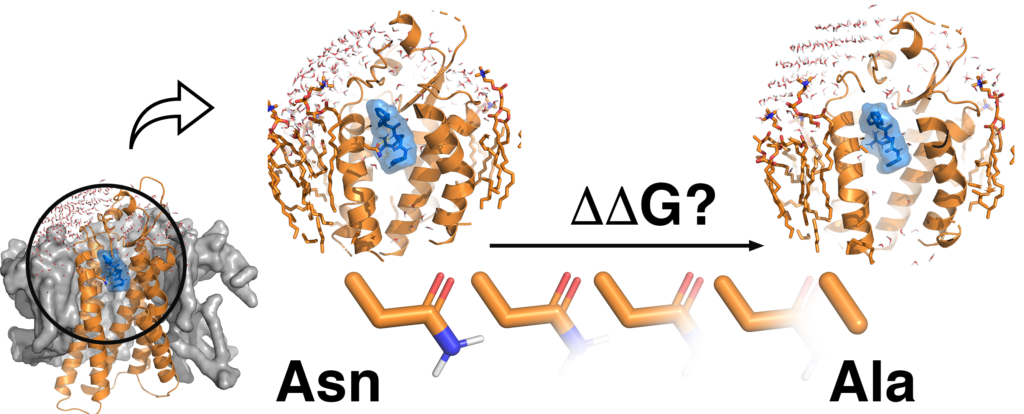
QresFEP: an automated protocol for free energy calculations of protein mutations in Q
W. Jespers, G.V. Isaksen, T.A.H. Andberg, S. Vasile, A. van Veen, J. Åqvist, B.O. Brandsdal, H. Gutiérrez-de-Terán, Journal of Chemical Theory and Computation 2019, 15 (10), 5461–5473.